My current scientific projects:
 The Origin of Runx Genes
The Origin of Runx Genes
How are genes “born” in evolution?
Tools: The Nematostella model and computational biology
A new study explores the evolutionary origins of the Runx genes, which are a metazoan novelty. For this aim, we have introduced the sea-anemone Nematostella vectensis, which is a new model organism representing a basal metazoan. We are studying the primordial Runx gene in this animal and aim to uncover its emergence in evolution.
 The Hippo Pathway of Organ Growth Regulation: Through an Evolutionary Looking Glass
What new insights can we get from an evolutionary stand point?
Tools: bioinformatics and computational biology. The sea-anemone Nematostella model system
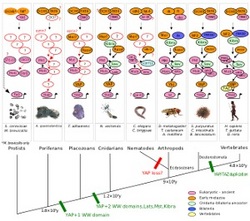
The Hippo pathway is an important and interesting developmental signaling pathway which plays an important role in animal organ size control. The pathway which is the latest major developmental pathway that was defined regulates tissue proliferation and apoptosis rates as a response to developmental cues, cell contact and density. We study the evolution of the Hippo pathway focusing on the transcriptional co-activator YAP, a pivotal effector of the pathway and an oncogene in human. We found most of the known mammalian components in the sea-anemone Nematostella vectensis, which displays a pre-bilaterian basal form of the pathway. Most of the major domains of YAP have been conserved between cnidarians and mammals. This project aims to study how the Hippo pathway operates in Nematostella and what can we learn from this basal animal about the general principals of its activity and how it has evolved.
The Hippo Pathway of Organ Growth Regulation: Through an Evolutionary Looking Glass
What new insights can we get from an evolutionary stand point?
Tools: bioinformatics and computational biology. The sea-anemone Nematostella model system
The Hippo pathway is an important and interesting developmental signaling pathway which plays an important role in animal organ size control. The pathway which is the latest major developmental pathway that was defined regulates tissue proliferation and apoptosis rates as a response to developmental cues, cell contact and density. We study the evolution of the Hippo pathway focusing on the transcriptional co-activator YAP, a pivotal effector of the pathway and an oncogene in human. We found most of the known mammalian components in the sea-anemone Nematostella vectensis, which displays a pre-bilaterian basal form of the pathway. Most of the major domains of YAP have been conserved between cnidarians and mammals. This project aims to study how the Hippo pathway operates in Nematostella and what can we learn from this basal animal about the general principals of its activity and how it has evolved.
Scientific publications and presentations
Gaffney CJ, Oka T, Mazack V, Hilman D, Gat U, Muramatsu T, Inazawa J, Golden
A, Carey DJ, Farooq A, Tromp G, Sudol M. Identification, basic characterization
and evolutionary analysis of differentially spliced mRNA isoforms of human YAP1
gene. Gene. 2012 Nov 10;509(2):215-22. doi: 10.1016/j.gene.2012.08.025. Epub 2012
Aug 24. PubMed PMID: 22939869; PubMed Central PMCID: PMC3455135.
Yakir E, Hassidim M, Melamed-Book N, Hilman D, Kron I, Green RM. Cell
autonomous and cell-type specific circadian rhythms in Arabidopsis. Plant J. 2011
Nov;68(3):520-31. doi: 10.1111/j.1365-313X.2011.04707.x. Epub 2011 Sep 26. PubMedPMID: 21781194.
Hilman D, Gat U. The evolutionary history of YAP and the hippo/YAP pathway.
Mol Biol Evol. 2011 Aug;28(8):2403-17. doi: 10.1093/molbev/msr065. Epub 2011 Mar
16. PubMed PMID: 21415026.
Yakir E, Hilman D, Kron I, Hassidim M, Melamed-Book N, Green RM.
Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian
oscillator of Arabidopsis. Plant Physiol. 2009 Jun;150(2):844-57. doi:
10.1104/pp.109.137414. Epub 2009 Apr 1. PubMed PMID: 19339503; PubMed Central
PMCID: PMC2689986.
Yakir E, Hilman D, Hassidim M, Green RM. CIRCADIAN CLOCK ASSOCIATED1
transcript stability and the entrainment of the circadian clock in Arabidopsis.
Plant Physiol. 2007 Nov;145(3):925-32. Epub 2007 Sep 14. PubMed PMID: 17873091;
PubMed Central PMCID: PMC2048808.
Hassidim M, Yakir E, Fradkin D, Hilman D, Kron I, Keren N, Harir Y, Yerushalmi
S, Green RM. Mutations in CHLOROPLAST RNA BINDING provide evidence for the
involvement of the chloroplast in the regulation of the circadian clock in
Arabidopsis. Plant J. 2007 Aug;51(4):551-62. Epub 2007 Jul 7. PubMed PMID:17617174.
Yakir E, Hilman D, Harir Y, Green RM. Regulation of output from the plant
circadian clock. FEBS J. 2007 Jan;274(2):335-45. Review. PubMed PMID: 17229141.